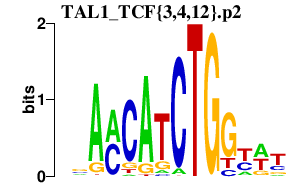
|
chr7_-_133591146
|
16.907
|
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr11_+_101192080
|
13.233
|
|
Aoc3
|
amine oxidase, copper containing 3
|
|
chr5_-_17394776
|
12.319
|
NM_001159558
|
Cd36
|
CD36 antigen
|
|
chr7_+_31561419
|
12.291
|
NM_028618
|
Dmkn
|
dermokine
|
|
chr11_-_5703752
|
11.779
|
NM_018870
|
Pgam2
|
phosphoglycerate mutase 2
|
|
chr18_+_44122894
|
10.919
|
NM_001081180
|
Spink5
|
serine peptidase inhibitor, Kazal type 5
|
|
chr2_+_35159780
|
10.867
|
|
Gsn
|
gelsolin
|
|
chr2_+_158328394
|
10.066
|
|
Adig
|
adipogenin
|
|
chr1_+_196765124
|
10.030
|
|
Cd34
|
CD34 antigen
|
|
chr1_+_196765014
|
9.384
|
NM_001111059
NM_133654
|
Cd34
|
CD34 antigen
|
|
chr1_+_196765079
|
8.716
|
|
Cd34
|
CD34 antigen
|
|
chr2_+_158328317
|
8.336
|
NM_145635
|
Adig
|
adipogenin
|
|
chr11_-_5703677
|
7.849
|
|
Pgam2
|
phosphoglycerate mutase 2
|
|
chr8_-_109589522
|
6.679
|
NM_025458
|
Tmed6
|
transmembrane emp24 protein transport domain containing 6
|
|
chr5_-_17394564
|
6.637
|
|
Cd36
|
CD36 antigen
|
|
chr7_+_28856254
|
6.361
|
NM_001122603
|
Fcgbp
|
Fc fragment of IgG binding protein
|
|
chr14_+_120674921
|
6.351
|
|
Mbnl2
|
muscleblind-like 2
|
|
chr14_+_120674886
|
6.272
|
NM_175341
NM_207515
|
Mbnl2
|
muscleblind-like 2
|
|
chr14_+_120674933
|
6.269
|
|
Mbnl2
|
muscleblind-like 2
|
|
chr2_-_79969095
|
5.582
|
NM_016744
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr4_-_148829073
|
5.043
|
NM_022020
|
Rbp7
|
retinol binding protein 7, cellular
|
|
chr14_+_120674913
|
4.996
|
|
Mbnl2
|
muscleblind-like 2
|
|
chr8_-_126417352
|
4.929
|
|
Acta1
|
actin, alpha 1, skeletal muscle
|
|
chr9_-_15105864
|
4.690
|
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene
|
|
chr14_+_120674939
|
4.419
|
|
Mbnl2
|
muscleblind-like 2
|
|
chr1_+_45402907
|
4.318
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr8_-_11199802
|
4.305
|
|
Col4a1
|
collagen, type IV, alpha 1
|
|
chr9_-_15105965
|
4.249
|
NM_133732
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene
|
|
chr2_-_122436933
|
4.201
|
NM_025961
|
Gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr10_-_75106702
|
4.104
|
NM_001162913
NM_027890
|
Susd2
|
sushi domain containing 2
|
|
chr15_+_58770843
|
3.480
|
|
Ndufb9
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9
|
|
chr3_-_8964045
|
3.479
|
NM_001025261
NM_001025262
|
Tpd52
|
tumor protein D52
|
|
chr18_-_34738725
|
3.469
|
NM_080637
|
Nme5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr1_-_164796609
|
3.450
|
|
Fmo1
|
flavin containing monooxygenase 1
|
|
chr17_+_24864773
|
3.411
|
NM_001163945
|
Rpl3l
|
ribosomal protein L3-like
|
|
chrX_+_68809128
|
3.355
|
|
Hmgb3
|
high mobility group box 3
|
|
chr1_-_164796660
|
3.164
|
NM_010231
|
Fmo1
|
flavin containing monooxygenase 1
|
|
chr11_+_67611772
|
3.125
|
NM_001013013
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C
|
|
chr5_-_44485951
|
3.091
|
NM_001163578
NM_001163581
NM_001163582
NM_001163583
NM_001163584
NM_001163585
|
Prom1
|
prominin 1
|
|
chr5_+_130263548
|
3.059
|
|
Gbas
|
glioblastoma amplified sequence
|
|
chr9_+_114373337
|
2.944
|
|
Glb1
|
galactosidase, beta 1
|
|
chr7_+_147949637
|
2.938
|
|
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1
|
|
chr3_+_19869841
|
2.927
|
|
Cp
|
ceruloplasmin
|
|
chr4_+_118199847
|
2.886
|
NM_025572
|
2610528J11Rik
|
RIKEN cDNA 2610528J11 gene
|
|
chr7_+_51480806
|
2.866
|
NM_010639
|
Klk1
|
kallikrein 1
|
|
chr11_+_96205082
|
2.844
|
NM_010458
|
Hoxb3
|
homeobox B3
|
|
chr11_+_93905501
|
2.774
|
NM_001025428
NM_001025429
NM_001025430
|
Spag9
|
sperm associated antigen 9
|
|
chr15_+_78859949
|
2.770
|
|
|
|
|
chr14_-_35401845
|
2.741
|
NM_001039071
NM_001039072
NM_001039073
NM_001039075
NM_001039076
NM_011918
|
Ldb3
|
LIM domain binding 3
|
|
chr1_-_195090178
|
2.726
|
NM_008288
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1
|
|
chr7_+_138087002
|
2.542
|
|
Htra1
|
HtrA serine peptidase 1
|
|
chr6_-_38787222
|
2.540
|
NM_001136065
|
Hipk2
|
homeodomain interacting protein kinase 2
|
|
chr4_+_148913978
|
2.523
|
|
Ctnnbip1
|
catenin beta interacting protein 1
|
|
chr17_+_75578120
|
2.517
|
NM_206958
|
Ltbp1
|
latent transforming growth factor beta binding protein 1
|
|
chr4_+_109088251
|
2.489
|
NM_001145948
|
Ttc39a
|
tetratricopeptide repeat domain 39A
|
|
chr7_+_27904997
|
2.422
|
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2
|
|
chr11_+_102697609
|
2.412
|
NM_080846
|
Higd1b
|
HIG1 domain family, member 1B
|
|
chr11_+_83560497
|
2.228
|
|
1100001G20Rik
|
RIKEN cDNA 1100001G20 gene
|
|
chr11_+_96790581
|
2.225
|
NM_146026
|
Prr15l
|
proline rich 15-like
|
|
chr1_+_133534852
|
2.212
|
NM_007799
|
Ctse
|
cathepsin E
|
|
chr6_+_5340417
|
2.181
|
|
Asb4
|
ankyrin repeat and SOCS box-containing 4
|
|
chr15_+_78860043
|
2.168
|
|
H1f0
|
H1 histone family, member 0
|
|
chr7_+_28902406
|
2.139
|
|
Fcgbp
|
Fc fragment of IgG binding protein
|
|
chr7_+_28914482
|
2.138
|
NM_001164655
|
9530053A07Rik
|
RIKEN cDNA 9530053A07 gene
|
|
chr1_+_34356614
|
2.138
|
|
Dst
|
dystonin
|
|
chr14_-_104213981
|
2.087
|
|
|
|
|
chr17_-_11033034
|
2.053
|
NM_027032
|
Pacrg
|
PARK2 co-regulated
|
|
chr12_-_102988596
|
2.052
|
|
|
|
|
chrX_+_68809103
|
2.045
|
|
Hmgb3
|
high mobility group box 3
|
|
chr16_+_16213397
|
2.038
|
|
Pkp2
|
plakophilin 2
|
|
chr11_+_93905443
|
2.021
|
|
Spag9
|
sperm associated antigen 9
|
|
chr6_+_122902850
|
1.960
|
|
Clec4a3
|
C-type lectin domain family 4, member a3
|
|
chr4_+_80556574
|
1.941
|
NM_026821
|
D4Bwg0951e
|
DNA segment, Chr 4, Brigham & Women's Genetics 0951 expressed
|
|
chr11_+_83560437
|
1.930
|
NM_183249
|
1100001G20Rik
|
RIKEN cDNA 1100001G20 gene
|
|
chr6_+_122902755
|
1.896
|
|
Clec4a3
|
C-type lectin domain family 4, member a3
|
|
chr11_+_29592897
|
1.855
|
NM_194052
NM_194053
NM_194054
|
Rtn4
|
reticulon 4
|
|
chr6_+_4471231
|
1.835
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr9_+_77961211
|
1.827
|
|
Ick
|
intestinal cell kinase
|
|
chr1_+_90034940
|
1.784
|
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A
|
|
chr4_-_40883509
|
1.727
|
|
Bag1
|
BCL2-associated athanogene 1
|
|
chr6_+_122902532
|
1.713
|
NM_153197
|
Clec4a3
|
C-type lectin domain family 4, member a3
|
|
chr10_-_87937335
|
1.710
|
|
Chpt1
|
choline phosphotransferase 1
|
|
chr3_+_108163704
|
1.696
|
|
Sort1
|
sortilin 1
|
|
chr1_+_132628552
|
1.679
|
NM_177604
|
AA986860
|
expressed sequence AA986860
|
|
chr4_+_94405963
|
1.653
|
NM_013690
|
Tek
|
endothelial-specific receptor tyrosine kinase
|
|
chrX_+_68809092
|
1.625
|
|
Hmgb3
|
high mobility group box 3
|
|
chr9_-_66891535
|
1.623
|
NM_001164252
NM_001164253
NM_001164254
NM_001164256
|
Tpm1
|
tropomyosin 1, alpha
|
|
chr11_+_120537974
|
1.605
|
NM_001164224
NM_198223
|
Aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 (human)
|
|
chrX_-_100958883
|
1.602
|
NM_001170979
|
Gm6222
|
predicted pseudogene 6222
|
|
chr17_-_28536156
|
1.590
|
|
Fkbp5
|
FK506 binding protein 5
|
|
chr7_-_26262552
|
1.589
|
NM_001039185
NM_001039186
NM_001039187
NM_011926
|
Ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1
|
|
chr11_-_3817574
|
1.566
|
|
Tcn2
|
transcobalamin 2
|
|
chr19_-_40525876
|
1.498
|
NM_001034962
NM_001034963
|
Sorbs1
|
sorbin and SH3 domain containing 1
|
|
chr15_+_78860623
|
1.480
|
|
|
|
|
chr3_+_101949321
|
1.421
|
|
Casq2
|
calsequestrin 2
|
|
chr3_-_95110715
|
1.394
|
NM_001085383
|
Anxa9
|
annexin A9
|
|
chr18_-_12978106
|
1.381
|
|
Osbpl1a
|
oxysterol binding protein-like 1A
|
|
chr5_+_23907911
|
1.371
|
|
Abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8
|
|
chr1_+_137732961
|
1.365
|
NM_001130174
NM_001130175
NM_001130177
NM_001130178
NM_001130179
NM_001130180
NM_001130181
NM_011619
NM_001136083
|
Tnnt2
|
troponin T2, cardiac
|
|
chr17_-_45724536
|
1.329
|
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr15_-_89307854
|
1.277
|
NM_009713
|
Arsa
|
arylsulfatase A
|
|
chr3_+_93123668
|
1.275
|
NM_133698
|
Hrnr
|
hornerin
|
|
chr2_-_164008333
|
1.255
|
NM_145369
|
Wfdc5
|
WAP four-disulfide core domain 5
|
|
chr16_+_18052952
|
1.243
|
NM_010047
|
Dgcr6
|
DiGeorge syndrome critical region gene 6
|
|
chrX_+_53708015
|
1.233
|
NM_172779
|
Ddx26b
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B
|
|
chr7_-_26324847
|
1.174
|
NM_001113368
NM_001113369
NM_007543
|
Ceacam2
|
carcinoembryonic antigen-related cell adhesion molecule 2
|
|
chr7_+_25378887
|
1.150
|
|
Ethe1
|
ethylmalonic encephalopathy 1
|
|
chr1_+_137732910
|
1.148
|
|
Tnnt2
|
troponin T2, cardiac
|
|
chr13_-_23677831
|
1.128
|
|
Hist1h2be
|
histone cluster 1, H2be
|
|
chr1_+_164569335
|
1.124
|
|
Myoc
|
myocilin
|
|
chr2_-_25477449
|
1.082
|
NM_001040130
NM_001109993
|
Tmem141
|
transmembrane protein 141
|
|
chr8_+_131028275
|
1.075
|
|
Nrp1
|
neuropilin 1
|
|
chr17_-_31281184
|
1.074
|
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1)
|
|
chr4_+_57867433
|
1.070
|
NM_001035533
|
Akap2
|
A kinase (PRKA) anchor protein 2
|
|
chr7_+_27904981
|
1.038
|
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2
|
|
chr1_-_71642303
|
1.010
|
|
Fn1
|
fibronectin 1
|
|
chr14_+_56236989
|
0.995
|
NM_020002
|
Rec8
|
REC8 homolog (yeast)
|
|
chr6_-_40985505
|
0.988
|
NM_023333
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene
|
|
chr3_-_95111053
|
0.986
|
NM_023628
|
Anxa9
|
annexin A9
|
|
chr16_-_31314534
|
0.984
|
NM_007470
|
Apod
|
apolipoprotein D
|
|
chr9_+_32031833
|
0.970
|
NM_177379
|
Arhgap32
|
Rho GTPase activating protein 32
|
|
chr7_-_147288242
|
0.960
|
|
1810014F10Rik
|
RIKEN cDNA 1810014F10 gene
|
|
chr3_-_126701367
|
0.954
|
NM_178655
|
Ank2
|
ankyrin 2, brain
|
|
chr3_-_105746740
|
0.953
|
|
Atp5f1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1
|
|
chr12_-_102988510
|
0.950
|
|
Fbln5
|
fibulin 5
|
|
chr19_-_23522811
|
0.937
|
NM_174857
|
Mamdc2
|
MAM domain containing 2
|
|
chr1_+_97230610
|
0.933
|
|
Fam174a
|
family with sequence similarity 174, member A
|
|
chr2_+_25036524
|
0.898
|
|
Nrarp
|
Notch-regulated ankyrin repeat protein
|
|
chr7_-_106844083
|
0.893
|
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1
|
|
chr8_-_71601126
|
0.890
|
|
Slc18a1
|
solute carrier family 18 (vesicular monoamine), member 1
|
|
chr11_-_70465629
|
0.889
|
|
Pfn1
|
profilin 1
|
|
chr17_-_34875879
|
0.885
|
|
C4b
|
complement component 4B (Childo blood group)
|
|
chr11_+_93905557
|
0.873
|
|
Spag9
|
sperm associated antigen 9
|
|
chr10_+_101621491
|
0.863
|
NM_001162368
|
Mgat4c
4930402I19Rik
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative)
RIKEN cDNA 4930402I19 gene
|
|
chr2_-_3339919
|
0.852
|
NM_008579
|
Meig1
|
meiosis expressed gene 1
|
|
chr7_+_20101391
|
0.825
|
|
|
|
|
chr4_-_116312481
|
0.824
|
|
Akr1a4
|
aldo-keto reductase family 1, member A4 (aldehyde reductase)
|
|
chr12_-_52847096
|
0.818
|
|
Hectd1
|
HECT domain containing 1
|
|
chr11_+_29592946
|
0.808
|
|
Rtn4
|
reticulon 4
|
|
chr1_-_141982814
|
0.803
|
|
Cfh
|
complement component factor h
|
|
chr7_-_150927867
|
0.793
|
NM_024289
|
Osbpl5
|
oxysterol binding protein-like 5
|
|
chr16_+_4741612
|
0.784
|
NM_010443
|
Hmox2
|
heme oxygenase (decycling) 2
|
|
chr6_-_3444484
|
0.771
|
NM_178899
|
Hepacam2
|
HEPACAM family member 2
|
|
chr10_+_60552061
|
0.769
|
NM_025273
|
Pcbd1
|
pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 1
|
|
chr1_-_172906094
|
0.764
|
NM_001077189
NM_010187
|
Fcgr2b
|
Fc receptor, IgG, low affinity IIb
|
|
chr2_+_174170886
|
0.742
|
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus
|
|
chr14_-_21437545
|
0.740
|
NM_030180
|
Usp54
|
ubiquitin specific peptidase 54
|
|
chr9_+_92502908
|
0.719
|
|
Plod2
|
procollagen lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr4_-_116312569
|
0.717
|
|
Akr1a4
|
aldo-keto reductase family 1, member A4 (aldehyde reductase)
|
|
chr9_-_110649625
|
0.716
|
NM_011199
|
Pth1r
|
parathyroid hormone 1 receptor
|
|
chr10_+_59747208
|
0.708
|
NM_001146124
|
Psap
|
prosaposin
|
|
chr4_-_40886191
|
0.707
|
|
Bag1
|
BCL2-associated athanogene 1
|
|
chr18_+_11997728
|
0.704
|
NM_001146287
NM_022021
|
Cables1
|
CDK5 and Abl enzyme substrate 1
|
|
chr17_-_31281223
|
0.699
|
NM_009363
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1)
|
|
chr14_+_103912557
|
0.697
|
NM_022886
|
Scel
|
sciellin
|
|
chr15_-_82169203
|
0.697
|
NM_008669
|
Naga
|
N-acetyl galactosaminidase, alpha
|
|
chr3_-_84063605
|
0.691
|
|
Trim2
|
tripartite motif-containing 2
|
|
chr10_+_18246228
|
0.678
|
|
Nhsl1
|
NHS-like 1
|
|
chr3_-_75073981
|
0.678
|
NM_026460
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2
|
|
chr1_-_36758359
|
0.663
|
|
Actr1b
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast)
|
|
chr8_-_73905790
|
0.648
|
NM_010150
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6
|
|
chr2_+_35413977
|
0.644
|
NM_001114125
|
Dab2ip
|
disabled homolog 2 (Drosophila) interacting protein
|
|
chr1_-_173177236
|
0.643
|
|
Ndufs2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2
|
|
chr7_+_72789752
|
0.643
|
NM_172310
|
Tarsl2
|
threonyl-tRNA synthetase-like 2
|
|
chr11_+_29593657
|
0.616
|
NM_194051
|
Rtn4
|
reticulon 4
|
|
chr3_+_95392919
|
0.615
|
|
Golph3l
|
golgi phosphoprotein 3-like
|
|
chr4_-_141154138
|
0.613
|
|
Fblim1
|
filamin binding LIM protein 1
|
|
chr1_-_195068455
|
0.601
|
NM_001044751
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1
|
|
chrX_+_147024463
|
0.599
|
NM_008824
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1
|
|
chr4_+_115761660
|
0.595
|
|
Lrrc41
|
leucine rich repeat containing 41
|
|
chr19_+_7495023
|
0.590
|
|
2700081O15Rik
|
RIKEN cDNA 2700081O15 gene
|
|
chr2_+_101726413
|
0.589
|
NM_029635
|
Commd9
|
COMM domain containing 9
|
|
chr2_+_31804066
|
0.589
|
|
1110064A23Rik
|
RIKEN cDNA 1110064A23 gene
|
|
chr16_-_18407565
|
0.575
|
|
Comt
|
catechol-O-methyltransferase
|
|
chr6_+_70676803
|
0.567
|
|
|
|
|
chr4_+_115769086
|
0.566
|
|
Lrrc41
|
leucine rich repeat containing 41
|
|
chr7_-_148462649
|
0.565
|
NM_001114322
NM_028069
|
Cdhr5
|
cadherin-related family member 5
|
|
chr19_+_34175422
|
0.552
|
NM_023903
|
Lipm
|
lipase, family member M
|
|
chr3_+_95392855
|
0.541
|
NM_001177669
NM_001177670
NM_146133
|
Golph3l
|
golgi phosphoprotein 3-like
|
|
chr12_-_86682028
|
0.541
|
|
|
|
|
chr4_+_139204697
|
0.538
|
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1
|
|
chr17_+_37035062
|
0.536
|
NM_146077
|
Trim31
|
tripartite motif-containing 31
|
|
chr4_+_98860586
|
0.534
|
NM_175029
|
Atg4c
|
autophagy-related 4C (yeast)
|
|
chr5_+_123526279
|
0.522
|
NM_001039723
|
Tmem120b
|
transmembrane protein 120B
|
|
chr17_-_26335980
|
0.514
|
NM_001081070
|
Pdia2
|
protein disulfide isomerase associated 2
|
|
chr7_-_149865451
|
0.511
|
NM_001185083
NM_001185084
NM_008387
|
Ins2
|
insulin II
|
|
chr2_-_32111045
|
0.511
|
|
Uck1
|
uridine-cytidine kinase 1
|
|
chr4_+_53838895
|
0.510
|
NM_028053
|
Tmem38b
|
transmembrane protein 38B
|
|
chr5_+_3236388
|
0.504
|
|
Gm15772
|
ribosomal protein L26 pseudogene
|
|
chr9_-_48413097
|
0.499
|
|
Nnmt
|
nicotinamide N-methyltransferase
|
|
chr9_+_78078473
|
0.483
|
NM_008181
|
Gsta1
|
glutathione S-transferase, alpha 1 (Ya)
|
|
chr5_+_31179203
|
0.483
|
|
Tmem214
|
transmembrane protein 214
|
|
chr7_-_135137189
|
0.480
|
|
Pycard
|
PYD and CARD domain containing
|
|
chr3_-_88058593
|
0.476
|
NM_021375
|
Rhbg
|
Rhesus blood group-associated B glycoprotein
|
|
chr19_-_32285445
|
0.471
|
NM_001168526
|
Sgms1
|
sphingomyelin synthase 1
|
|
chr9_+_78078492
|
0.459
|
|
Gsta1
|
glutathione S-transferase, alpha 1 (Ya)
|
|
chr13_-_61671425
|
0.458
|
NM_026906
|
Cts3
|
cathepsin 3
|
|
chr7_-_125676823
|
0.450
|
NM_009940
|
Coq7
|
demethyl-Q 7
|
|
chr8_+_74662546
|
0.447
|
|
Tpm4
|
tropomyosin 4
|
|
chr7_-_126602780
|
0.446
|
NM_025989
|
Gp2
|
glycoprotein 2 (zymogen granule membrane)
|